DEMO 3: Shape from Defocus via Diffusion
Shape from Defocus via Diffusion
Demonstration of the Shape from Defocus via Diffusion method
clear all close all
Data set initialization
set the maximum number of iterations
MaxIterations = 100; % load input images and initialize parameters load('DataSet.mat'); % first resize scalingFactor = 1/2; I1 = imresize(I1,scalingFactor); % limit number of computations I2 = imresize(I2,scalingFactor); % limit number of computations % select region of interest I(:,:,2) = I1(1:end-10,60:end-50); I(:,:,1) = I2(1:end-10,60:end-50); [M,N,K] = size(I); % lens parameters F = 12e-3; % focal length Fnumb = 2; % F number D = F/Fnumb; % aperture (effective lens diameter) gamma = 3e4; % calibration parameter p = 1./(1/F-1./[.52 .85]); % distance CCD-lens) % set camera parameters parms.m = M; parms.n = N; parms.F = F; parms.Fnumb = Fnumb; parms.D = D; parms.gamma = gamma; % same settings for second image parms(2) = parms(1); % plane in focus distance (from the lens) parms(1).p = p(1); parms(2).p = p(2); % select the domain where the data term is considered ww = 3; mask = ones(M,N); mask(1:ww,:) = 0; mask(end-ww+1:end,:) = 0; mask(:,1:ww) = 0; mask(:,end-ww+1:end) = 0; dt_data = 2e-1;%6e-1; % data term step dt_reg = 1e0;%2e0; % regularization term step maxDelta = 1; % max amount of relative blur win = 5; % regularization of initial depth map K = length(p); % number of focus settings thresh = 6e-2;% preconditioning threshold
Initialize unknown shape
initialize depth map with a plane
DepthMap_e = sum(p)*F/(sum(p)-2*F)*ones(M,N);
% compute the relative diffusion map
Sigma1 = (gamma*D/2*abs(1-p(1).*(1/F-1./DepthMap_e))).^2;
Sigma2 = (gamma*D/2*abs(1-p(2).*(1/F-1./DepthMap_e))).^2;
Dsigma = Sigma2-Sigma1;
Start shape estimation
integrate the gradient-flow for MaxIterations iterations
for i=1:MaxIterations % Optimization procedure starts here modelnoise = 0; sensornoise = 0; Omega = double(Dsigma>0); % Heaviside function delta = zerocrossing(Dsigma); % Dirac delta % compute diffusion PDE solution by simulation [dummy,u1] = synthesize(I(:,:,1),Dsigma.*Omega,... modelnoise,sensornoise,5); [dummy,u2] = synthesize(I(:,:,2),-Dsigma.*(1-Omega),... modelnoise,sensornoise,5); % compute initial conditions for the adjoint PDE w01 = reshape(u1(:,:,end),M,N)-I(:,:,2); % compute adjoint diffusion PDE solution by simulation [dummy,w1] = synthesize(w01,Dsigma.*Omega,... modelnoise,sensornoise,5); w02 = reshape(u2(:,:,end),M,N)-I(:,:,1); % compute adjoint diffusion PDE solution by simulation [dummy,w2] = synthesize(w02,-Dsigma.*(1-Omega),... modelnoise,sensornoise,5); % cost functional gradients computation % time integration (from 0 to T) of grad v grad u [dvdu1] = timeintegrate(u1,w1); % time integration (from 0 to T) of grad v grad u [dvdu2] = timeintegrate(u2,w2); dEdc = -2*Omega.*dvdu1+2*(1-Omega).*dvdu2+... delta.*((u1(:,:,end)-I(:,:,2)).^2-... (u2(:,:,end)-I(:,:,1)).^2); % preconditioning [dummy,p1] = synthesize(reshape(I(:,:,2),M,N,1),... Dsigma.*Omega,modelnoise,sensornoise,5); [dummy,p2] = synthesize(reshape(I(:,:,1),M,N,1),... -Dsigma.*(1-Omega),modelnoise,sensornoise,5); % time integration (from 0 to T) of grad p grad u [dpdu1] = timeintegrate(u1,p1); [dpdu2] = timeintegrate(u2,p2); precond = 1+abs(-2*Omega.*dpdu1+2*(1-Omega).*dpdu2); dEdc = dEdc./precond; dEdc = (abs(dEdc) < thresh).*dEdc+... (abs(dEdc) >= thresh).*thresh.*sign(dEdc); dDatads = dEdc.*mask; % make sure the number of iterations is an even number nIt = 2*(floor(dt_reg/1e-1)+1); beta_data = dt_data/nIt; beta_regularization = dt_reg/nIt; for tt=1:nIt % compute gradient of the cost functional (data term + % regularization) wrt the depth map if tt-floor(tt/2)*2==0 dsigma = beta_data*dDatads - beta_regularization.*... laplacian(Dsigma,'forward'); else dsigma = beta_data*dDatads - beta_regularization.*... laplacian(Dsigma,'backward'); end Dsigma = Dsigma-dsigma; end DepthMap_e = 1./(1/F-(p(2)-p(1)-sqrt((p(2)-p(1))^2+... 4*(p(2)^2-p(1)^2)*Dsigma/gamma^2/D^2))/... (p(2)^2-p(1)^2)); end
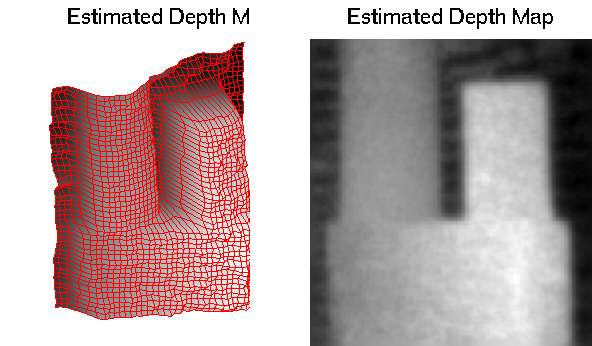
Results visualization
visualize input images and the estimated depth map (as a mesh and in gray-scale)
figure(1) subplot(211) image([I(:,:,1) 200*ones(size(I,1),10) ... I(:,:,2) 200*ones(size(I,1),10) ... 256*(DepthMap_e>sum(p)*F/(sum(p)-2*F))]) axis equal axis off set(gca,'XTick',[],'YTick',[]); set(gca,'Box','off'); colormap gray(256) title('Input #1/Input #2/Estimated Segmentation') drawnow; step = ceil(min([size(DepthMap_e,1) ... size(DepthMap_e,2)])/50); pix2mm = 1/600; [X,Y] = meshgrid(pix2mm*(1:step:size(DepthMap_e,2)),... pix2mm*(1:step:size(DepthMap_e,1))); axisZ = 1./(1/F-1./p); subplot(223) h = surf(X,-DepthMap_e(1:step:end,1:step:end),-Y,DepthMap_e(1:step:end,1:step:end)); shading interp hold off axis([0 N -axisZ(2) -axisZ(1) -M 0]) set(gca,'XTick',[],'YTick',[],'ZTick',[]); colormap gray(256) title('Estimated Depth Map') view([-1 -2 1]); axis equal axis off set(h,'EdgeColor','r'); drawnow; subplot(224) imagesc(DepthMap_e); colormap gray(256) axis equal set(gca,'XTick',[],'YTick',[]); set(gca,'Box','off'); title('Estimated Depth Map') drawnow;